Therapeutics & Nano-Pharmaceuticals
Parabon Essemblix™ Drug Development Platform
Key to the approach is the use of synthetic DNA as a programmable molecular substrate. Although DNA is best known as a carrier of genetic information, strands of synthetic DNA can be constructed to have any sequence of bases (often represented by the letters A, C, G and T).
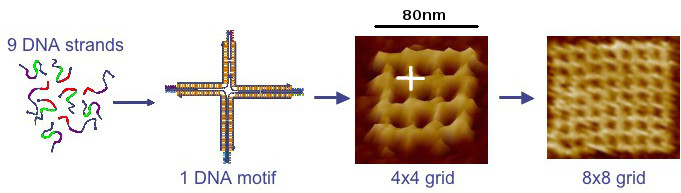
Because complementary sequences of DNA are mutually attractive, synthetic strands can be "programmed" with sequences that cause them to "swim to the right spot," with respect to one another, and then bind to form nanostructures of virtually any shape. Before self-assembly is induced, DNA strands can be attached to other types of molecular subcomponents so that they are pulled into designated locations by the DNA strands during self-assembly.
The Parabon inSēquio™ Design Studio
The inSēquio Design Studio allows Parabon's pharmaceutical engineers to graphically enter designs and then, using the extreme-scale computing capacity of Parabon's Frontier® Compute Platform, determines the optimal DNA sequences that will self-assemble into the specified design.
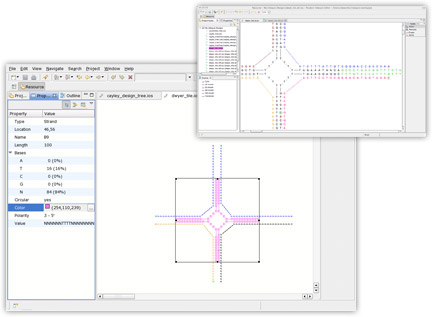
inSēquio's simple-to-use graphical editor allows nano-engineers to lay out a nanostructure visually. Users can rotate and bend strands, define bindings between base pairs, and copy and paste sequences and structures between design documents.
To learn more, or to explore partnership opportunities, please contact us.
DNA Phenotyping & Forensic Analysis
Parabon Snapshot® Advanced DNA Analysis Service
Recent advances in DNA sequencing technology have made it practical and affordable to read genetic content from DNA, which in turn has allowed the creation of datasets that include both genotypic (genetic content) and phenotypic (trait) data for each of thousands of subjects. With the diligent and repeated application of data mining and machine learning processes to such data, Parabon NanoLabs produces statistical models that translate the presence of specific genetic biomarkers into forensically relevant trait predictions.
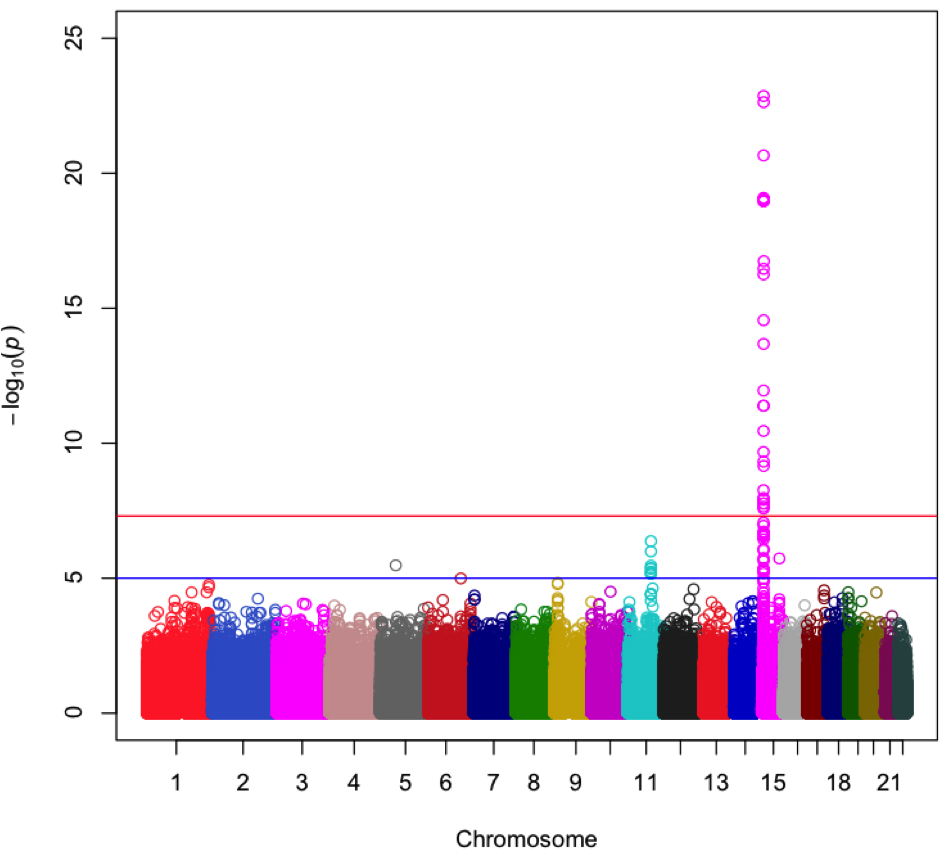
By mining genomic data, Snapshot identifies regions strongly associated with forensic traits. The above plot shows a region of chromosome 15 that is associated with eye color.
Beginning with large datasets comprised of a phenotype (trait) of interest and genotype data for thousands of subjects, our bioinformatics team performs large-scale statistical analysis on millions of individual SNPs and billions of combinations thereof to identify sets of these genetic markers that associate with the given trait. This mining process can take weeks running on hundreds, sometimes thousands, of computers. In the end, those SNPs with the greatest likelihood of contributing to the variation observed in the target trait are culled for potential use in predictive models.
The modeling phase further refines this set of SNPs to a final set that most accurately predict the target trait under a framework of machine learning algorithms. Models are validated against data held out for such testing and calibrated with all available data before being installed into the Snapshot architecture.


