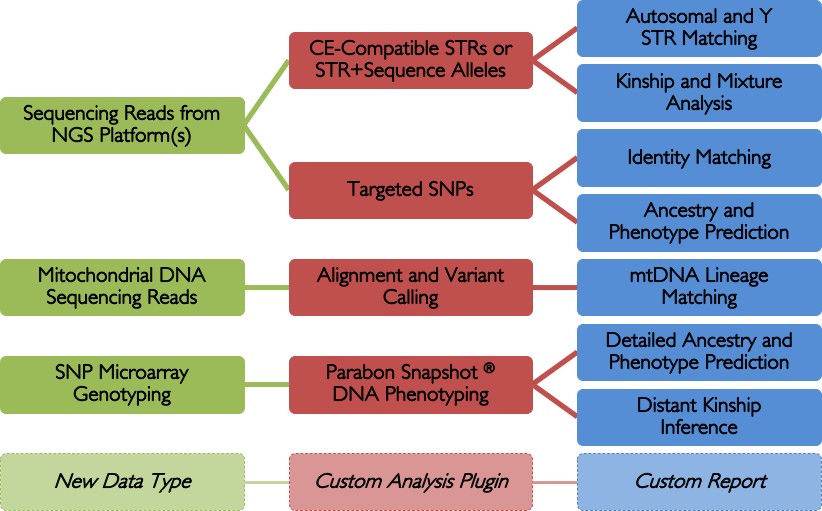
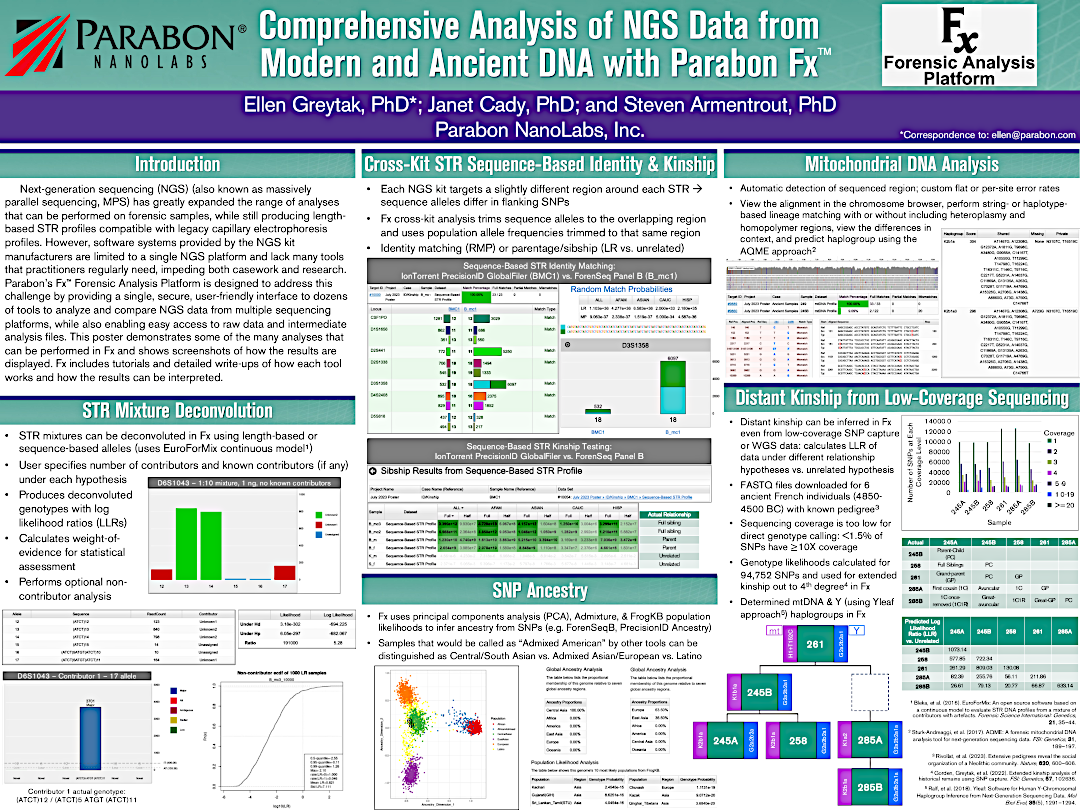
From targeted next-generation sequencing (NGS) for identity matching to whole-genome SNP microarray panels for ancestry and phenotype determination, the Fx platform offers a powerful yet user-friendly environment for importing, storing, analyzing and reporting on complex forensic datasets. Fx is vendor agnostic, allowing raw data to be imported from virtually any forensic instrument, and its extensible plugin architecture enables a wide collection of forensic apps to interoperate seamlessly under a consistent user interface.
- Transform raw data from multiple platforms and devices into meaningful results
- Perform complex analyses — such as ancestry, identity matching, STR sequence allele analysis, phenotype prediction, mixture deconvolution, and kinship inference — with the click of a button
- Track data provenance and audit usage
Powerful, Extensible, Cross-Platform
Analyze Data from STRs, SNPs, Mitochondrial DNA, and more. Next generation sequencing (NGS), also known as massively parallel sequencing (MPS) or high-throughput sequencing (HTS), is a powerful new forensic tool that that can analyze STRs, SNPs, and mitochondrial DNA from highly fragmented DNA samples, while providing greater discrimination among alleles by sequencing STRs rather than just calculating their length.
The Parabon Fx platform offers a single, unified interface to ingest data from any NGS system — e.g., Verogen ForenSeq, ThermoFisher Precision ID, Promega PowerSeq, etc. — and run an ever-increasing variety of analyses. Plus, it's extensible, so it can adapt to future kits as they become available.
STR Analysis
The Fx platform has been designed to process FASTQ files quickly and easily. Fx can call CE-compatible alleles for autosomal, X, and Y STRs from any sequencing instrument; call enhanced alleles containing sequence variants in the repeat region or flanking regions; flag loci with questionable allele calls; perform identity, kinship, and lineage comparisons to other samples using length- or sequence-based STR alleles, regardless of sequencing instrument; calculate RMP and LR; and perform mixture deconvolution.
Fx can also align and call mtDNA SNPs and perform string-based or haplotype-based lineage matching.
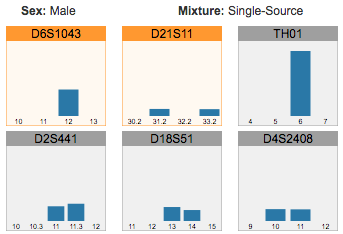
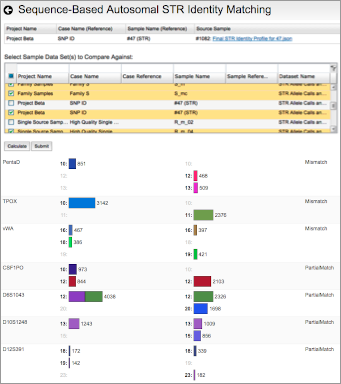
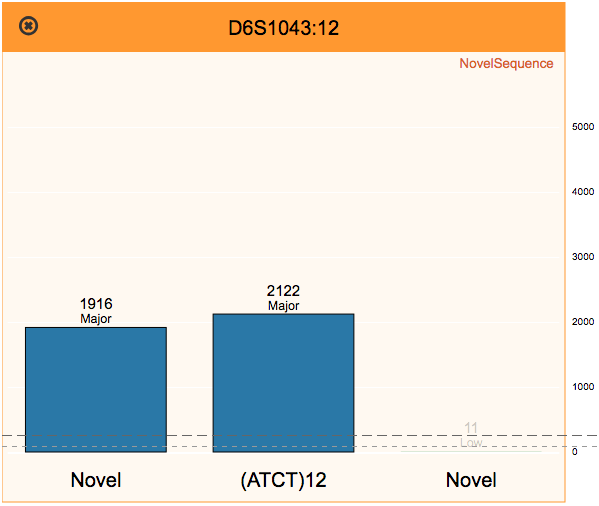
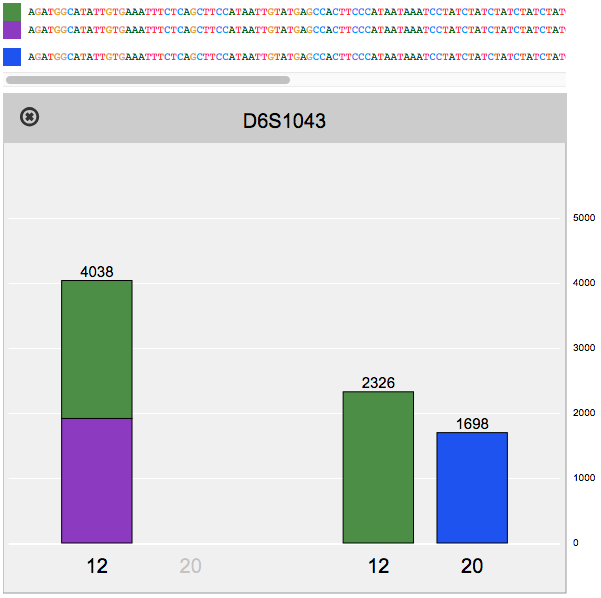
Reviewing flagged STR loci, using the finalized profile to perform identity matching, and comparing STR sequence alleles between two samples.
SNP Analysis
NGS data can also contain targeted SNPs, which can be obtained from samples too degraded for STRs. Fx can perform alignment and variant calling; display detailed read alignments using IGV; call and flag SNP alleles; perform identity matching; and predict ancestry and phenotype.
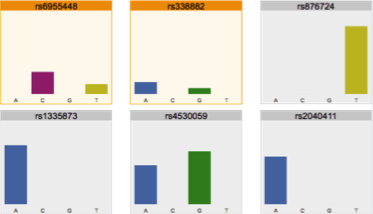
Reviewing flagged loci in an NGS
SNP Profile and predicting phenotype
In addition to NGS, the Fx platform can also ingest SNP microarray data. The hundreds of thousands of SNPs in such data can be used for in-depth analyses, such as comprehensive phenotyping, distant autosomal kinship inference, and precise ancestry determination.
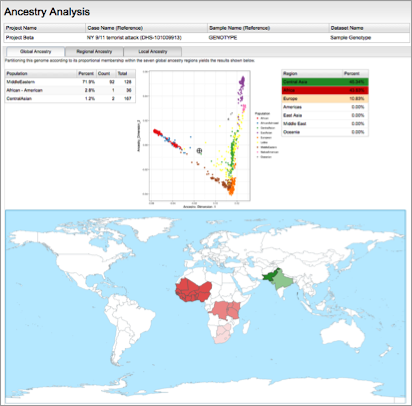
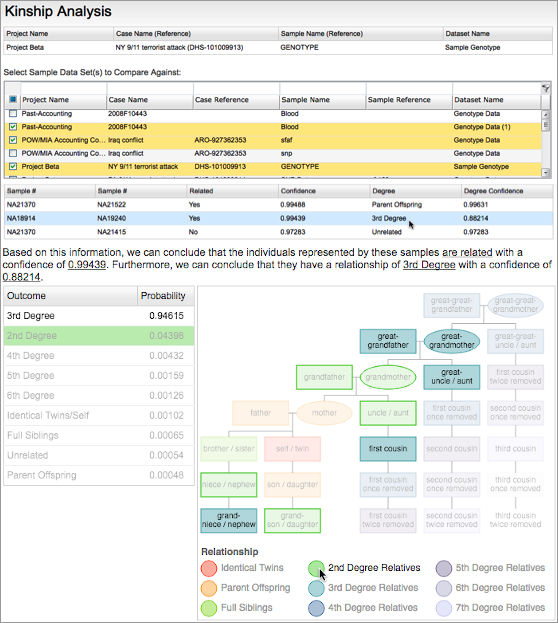
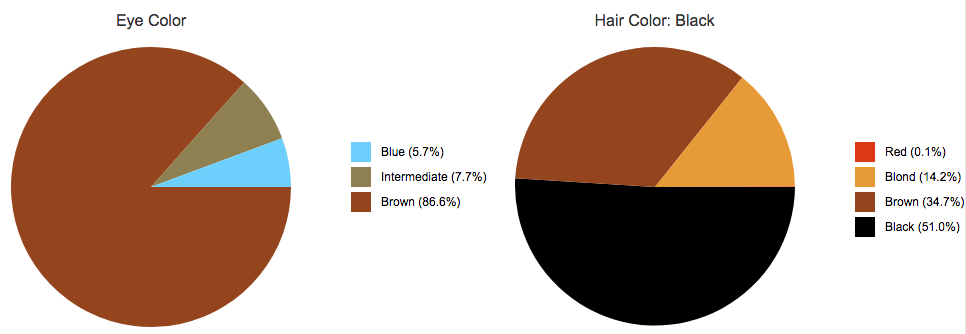
SNP microarray genotyping enables deep analyses, such as detailed ancestry (Left) and distant kinship inference (Right).
See What Fx Can Do
Development of the Fx platform is sponsored by the US Army Research Office. Funding is provided by the Department of Defense Office of the Deputy Assistant Secretary of Defense for Emerging Capabilities and Prototyping. This support does not imply a product endorsement from either entity.
For more information, please email fx-sales@parabon.com or call (703) 689-9689 x207


